There was this very embarrassing day around year six of my career as a statistician working in clinical trials. I had a small group of interns working on a project that combined data from multiple clinical trials. The goal was to better understand sources of variation in the common control used in a series of trials. Pretty routine stuff.
What made this bad for me was when I was reviewing a block of code. I got very aggravated with trying to understand what it was doing and why “they” chose to do it this particular way. The very nice interns did not act defensive at all. I finally exclaimed: “Where did you get the idea for the awful approach, some crazy Google search?” One of the interns very hesitantly said they pulled it from the source code I had given them. It turns out this same block was used in each of the entire sequence of trials I had them working with. Who was the author of this block? Unfortunately, the answer was a younger and more careless version of me.
Is it just me?
It turns out this topic has a great name - reproducible research. This is not to be confused with “replicable research” where similar methods are applied to new data to verify similar outcomes. In reproducible research, the goal is for a new researcher to take the same raw data and code and exactly match the reported results without much effort. The key being the without much effort part. I’ll get to that in a minute.
The origin
This term became very popular after a 2006 study published in Nature Medicine was examined by Keith Baggerly at MD Anderson Cancer Center in an attempt to reproduce the results using the same data. In that case, the research was not only hard to reproduce but it had been biased by data manipulation that was uncovered in the years that followed.
The story was featured on the CBS news show 60 Minutes Deception at Duke: Fraud in Cancer Care?. There is a great overview of the story by Keith Baggerly himself.
Without much effort
Now back to the without much effort part of making my code reproducible. With every trial submission to the FDA, we provide data in a standard format (SDTM) and our code for analysis, tables, listings and figures. The known context of these data sources make our code easier to understand, but does it match the without–much-effort standard?
Back in 2006, I used my embarrassing example as motivation to do a few things better when I created code. First, liberal use of comments to say what I am doing, describe how I was doing it and to note any sources of information. Another practice I added was creating a readme.txt file in folders with easy-to-read notes about the analysis approach I was taking as well as any assumptions and interpretation that were important.
Finding a solution
As I moved further into managing groups of statisticians and programmers, it became more important to find ways for teams of people to adhere to these practices. With that also came the obligation to make it easy to do. I have continued to seek new ways to make this easy in my career at SAS. Below I will introduce three approaches that we will dig deeper into with future blog posts.
A platform approach
I highly recommend looking at is SAS life sciences analytics software recently covered in a blog post by my colleague Matt Becker. It has many features built-in that make reproducibility easier with minimal burden on the creator of content. It also enables collaboration in a unique way that facilitates reproducibility.
SAS® Studio
This is the most modern development environment for SAS coders and users. It is browser-based and resembles the layout many of us have used our whole careers with quick access to the code, log and listing. SAS Studio includes a number of features that are very helpful for reproducible research:
- Legibility: Make code easier to read.
- Reuse: Code snippet functionality.
- Summary: Program summary with code, log, results and execution information.
- Package: SAS program Package with code, log and results.
- Custom Tasks: Users can interact with your code without programming.
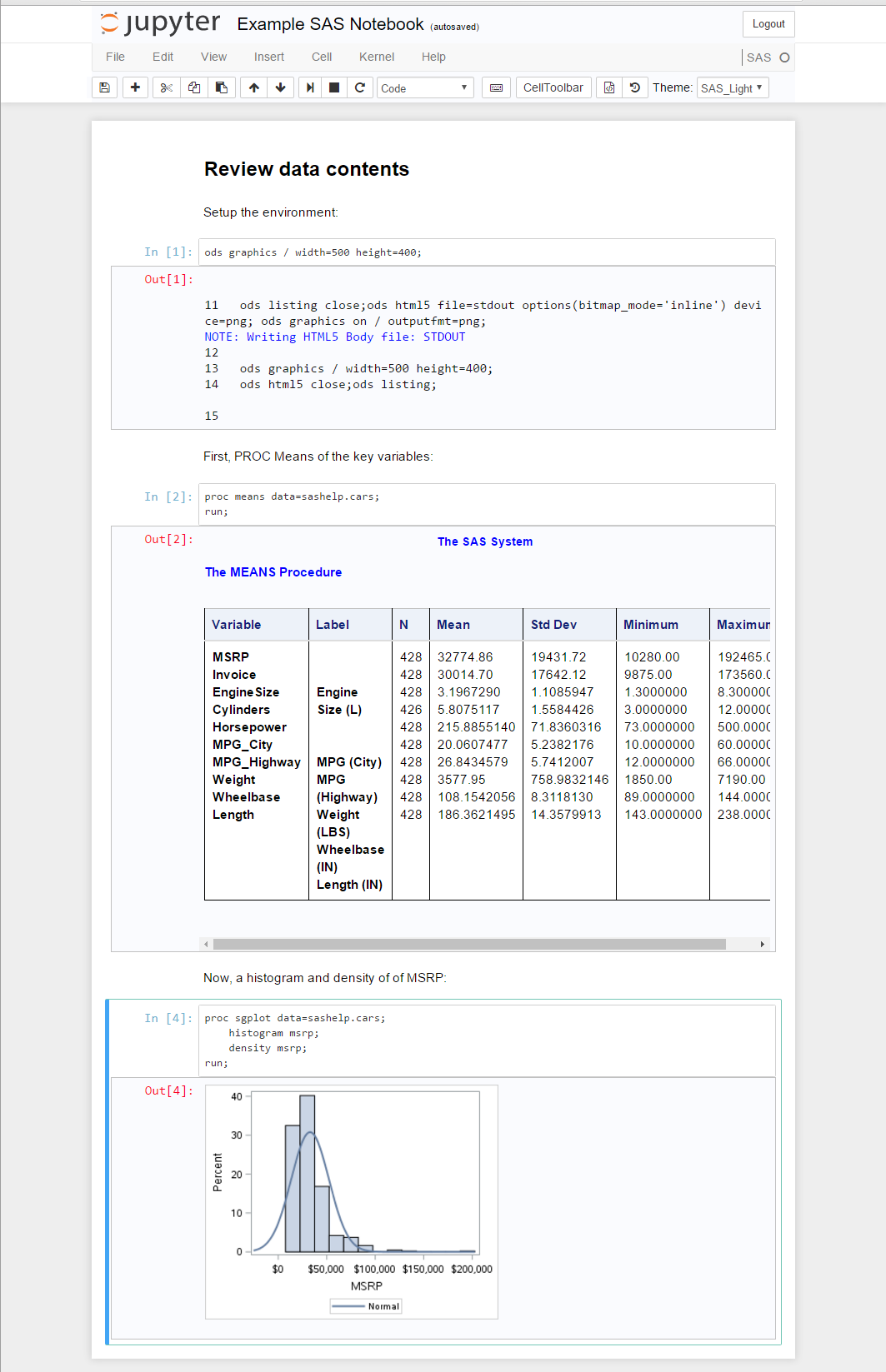
Notebooks
The notebook approach to coding involves embedding code into a virtual notebook that enables the creation of literate programming. A popular open-source notebook that is enabled by kernels to many languages, including SAS, is Jupyter. This allows the author to create very descriptive text alongside code blocks that can also be executed in line to display results in blocks. I like to describe this as a combined code, log and listing in a single document flow. A great introduction can be found in this blog post: How to run SAS programs in Jupyter Notebook
Let’s talk!
At the upcoming DIA 2016 Annual Meeting in Philadelphia, we will be demonstrating the three approaches introduced above. We would love to get your feedback. Please stop by Booth 1825 to see these ideas in action.

2 Comments
2006 is definitely not the origin of reproducible research; it was much earlier. Here is a 1995 paper on the topic by Buckheit and Donoho:
http://statistics.stanford.edu/sites/default/files/EFS%20NSF%20474.pdf
My paper discusses how user-written code can be released for reproducible research, but that system-generated code usually cannot be released, for copyright reasons:
http://www.wuss.org/proceedings14/5_Final_Paper_PDF.pdf
Thank you for this Thomas. This is a great addition to the post.
My initial intent for the section "The Origin" was to cover the origin but I found it difficult to trace the origin and opted to cover an inflection point in clinical research. Unfortunately, I did not rename the section. What are your thoughts on how RR can be achieved with SAS code today?
I also uncovered a system called the SAS StatRep System for Reproducible Research. I will cover this in a future post as well.