I sit here fascinated, watching a world population clock. According to the US Census, a baby is born and a person dies every 7 and 12 seconds, respectively, in the United States. In 1950 our world had about 2.5 billion people; currently our population is over 7.3 billion. Estimates predict over 9 billion people will be on this earth by 2050, and some experts believe that number to be conservative; estimating that a stabilization won’t happen for another 70 years, at around 12 billion.
How are we going to feed this many people?
One more large number is 3 billion: the estimated number of people (over 40%) that are currently malnourished. Technological/medical advances and new understanding of the human genome have improved both our quality of life and longevity. Now that same attention must be focused on our food sources.
What does this have to do with the software tools developed here at SAS? Well, researchers at places like the USDA and General Mills, to name just a couple, are using it to help solve this problem. Because grains and cereals make up over 80% of the world food sources they are in front-line position to help. They use JMP Genomics and SAS/OR to transform breeding programs with marker-assisted plant breeding.
The idea is to associate desirable traits in crops with locations on their genome so that we can help drive selection of new plant lines to optimize trait outcomes. A basic understanding of genetics is now common, especially with controversies surrounding genetically modified organisms (GMOs). If two blue-eyed people have a child, that child most likely will have blue eyes due to genetic code inherited from his/her parents. Cross two strains of corn that are tall, and the outcome will tend to be tall corn. These are called heritable traits. The goal then is to apply genomic knowledge of heritable traits to assist plant breeders in making the best crosses (mating of two plant lines) to produce crops that not only increase yield, but have less disease, have higher vitamin content, are easier to mill, can grow in drought-tolerant conditions, require less processing, and even taste better (yes, hedonic traits can and are being researched for crop improvement programs) without direct genetic modification.
How can our software help with this?
By developing tools that use the observed relationships between genetic markers to build a genetic map that facilitates trait association/prediction, crop selection, and simulation. Unlike the human genome, of which the majority has now been mapped, we don't have this “genetic roadmap” for many plant species due to strong complexities in their genome; for example, four, six, or even eight sets of chromosomes instead of two. Instead, we typically use the inheritance patterns between genetic markers to estimate a linkage map.
Markers inherited together are correlated, meaning they are closer on the genetic map. So we need algorithms that can group and order markers to build a genetic map. Markers that are correlated (a small genetic recombination distance) to a certain degree belong on the same chromosome. Once markers are assigned a chromosome, we need to determine the order of those markers that will produce the smallest genetic distance map.
It turns out this is directly analogous to the traveling salesman problem (TSP). Lucky for me, SAS just happens to have a group of experts in operations research that easily recognized this as an optimization problem using minimum spanning forests to determine groups of markers, and TSP algorithms in SAS/OR's OPTMODEL and OPTNET procedures to find optimal marker orders.
“Hey it’s not that simple! There are also known genetic relationships that might anchor certain markers to groups/orders,” I said; my new best friends in SAS/OR said, “No problem, we got this!” and applied optional node connection constraints in a side-constrained TSP solution.
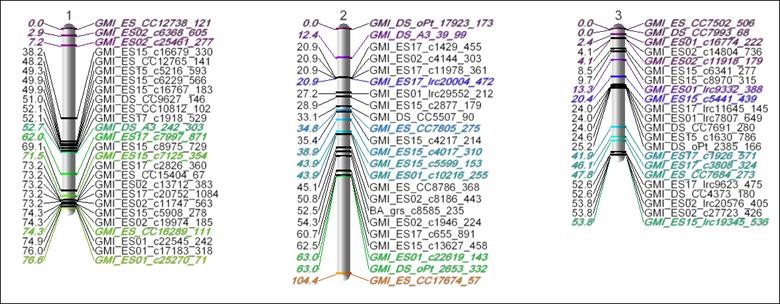
The Figure above shows an estimated Linkage Map using these methods for an experimental Oat population, where colored/italicized markers were already known/anchored markers. This optimization work and JMP Genomics visualization tools for linkage and consensus maps have since been used in several research publications for genetic mapping of a variety of plants, including this SAS Global Forum paper.
Predicting Traits, Cross Evaluation, and Breeding Program Simulation
With a reliable map, employing genomic selection methods (modern data mining and predictive modeling techniques for marker-assisted plant selection) becomes feasible in a plant breeding program. Using the arsenal of robust predictive models available in JMP Genomics, the goal is to find the best model to score a new plant variety for a given trait, based on genetic variability. The art of plant breeding comes in developing a new line (seed variety) that can balance multiple traits to give the best possible outcome. For example, just driving an increase in yield alone will often lead to the plant/product suffering in other ways.
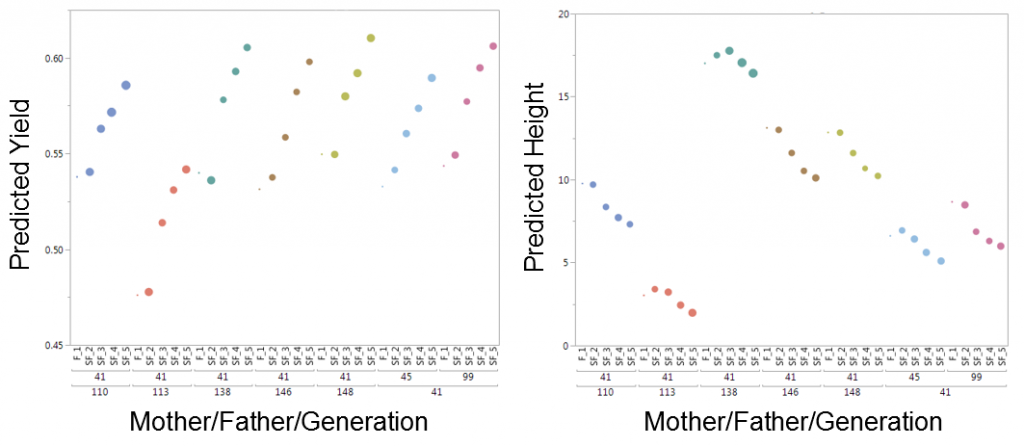
Our solution in JMP Genomics once again uses our secret weapon found in the capabilities of SAS/OR to create a process that allows breeders to analytically evaluate all possible plant/line crosses and create a cross simulation tool that selects the potential plant crosses that will simultaneously optimize multiple traits at once (for example in the plot below, increasing yield while decreasing height in the selected maize plant crosses).
The figure above shows that in five generations of breeding, crossing lines 45 or 99 with line 41 would produce some of the highest predicted yields while effectively reducing the height of the plant. A traditional breeding program can take several years before gain is realized. Each year, breeders have to decide the right crosses to make under the constraints of the amount of land/resources and time available to try to improve a given crop, and then must wait an entire growing season to see how the lines perform. JMP Genomics offers an analytic solution to help breeders find optimal crosses for a set of balanced physical traits and use multi-year simulation results to dramatically speed up this process, in many cases requiring just a few hours on a computer.
Creating sustainable agriculture techniques to produce not just more but healthier food is one of our most pressing concerns. The new methods outlined above provide just one way that SAS is able to help.